
This Processing Script is made available under the CC-0 license.
Fill nodata water pixels in DEM
from qgis import processing
from qgis.PyQt.QtCore import QCoreApplication, QVariant
from qgis.core import (
QgsProcessing,
QgsProcessingAlgorithm,
QgsProcessingParameterRasterLayer,
QgsProcessingParameterNumber,
QgsProcessingParameterFileDestination,
QgsProcessingContext
)
from osgeo import gdal, gdalconst
from pcraster import (
readmap,
ifthenelse,
defined,
boolean,
spreadmaxzone,
clump,
nominal,
cover,
areaminimum,
pcrnot,
report,
setglobaloption,
setclone
)
import os
import shutil
class FillWaterNoDataPixels(QgsProcessingAlgorithm):
INPUT_DEM = 'INPUT_DEM'
SIEVE_THRESHOLD = 'SIEVE_THRESHOLD'
OUTPUT_FILLED = 'OUTPUT_FILLED'
def tr(self, string):
return QCoreApplication.translate('Processing', string)
def createInstance(self):
return FillWaterNoDataPixels()
def name(self):
return 'fillnodatawaterpixels'
def displayName(self):
return self.tr('Fill nodata water pixels in DEM')
def group(self):
return self.tr('PCRaster User Scripts')
def groupId(self):
return 'pcrasteruser'
def shortHelpString(self):
return self.tr(
"""<html><body>
<p>Fill nodata water pixels in DEM</p>
<h2>Parameters</h2>
<b>DEM</b> (required) - DEM in raster format<br>
<b>Minimum sieve threshold</b> (required) - Minimum patch size (pixels) for sieving. 0 to skip sieving<br>
<b>Output water filled DEM</b> (required) - Output DEM in PCRaster format<br>
Author: Hans van der Kwast (QWAST-GIS)
</body></html>"""
)
def initAlgorithm(self, config=None):
self.addParameter(
QgsProcessingParameterRasterLayer(
self.INPUT_DEM,
'Input DEM'
)
)
self.addParameter(
QgsProcessingParameterNumber(
self.SIEVE_THRESHOLD,
'Minimum sieve threshold',
defaultValue=10,
minValue=0
)
)
self.addParameter(
QgsProcessingParameterFileDestination(
self.OUTPUT_FILLED,
'Output water filled DEM',
fileFilter='PCRaster map (*.map)'
)
)
def ConvertToPCRaster(self, src_filename, dst_filename, ot, VS):
# If source and destination are identical, skip conversion.
if os.path.abspath(src_filename) == os.path.abspath(dst_filename):
return
src_ds = gdal.Open(src_filename)
dst_ds = gdal.Translate(dst_filename, src_ds, format='PCRaster', outputType=ot, metadataOptions=VS)
dst_ds, src_ds = None, None
def sieve(self, src_filename, dst_filename, threshold):
if threshold > 0:
src_ds = gdal.Open(src_filename, gdal.GA_ReadOnly)
src_band = src_ds.GetRasterBand(1)
driver = gdal.GetDriverByName("GTiff")
dst_ds = driver.Create(dst_filename, src_ds.RasterXSize, src_ds.RasterYSize, 1, src_band.DataType)
dst_band = dst_ds.GetRasterBand(1)
gdal.SieveFilter(src_band, None, dst_band, threshold=threshold, connectedness=4)
dst_ds, src_ds = None, None
else:
# When sieve is skipped, simply copy the file.
if os.path.abspath(src_filename) != os.path.abspath(dst_filename):
shutil.copy(src_filename, dst_filename)
def processAlgorithm(self, parameters, context, feedback):
# Use QGIS's temporary folder for intermediate files.
temp_folder = context.temporaryFolder()
input_dem = self.parameterAsRasterLayer(parameters, self.INPUT_DEM, context).source()
sieve_threshold = self.parameterAsInt(parameters, self.SIEVE_THRESHOLD, context)
output_filled = self.parameterAsString(parameters, self.OUTPUT_FILLED, context)
# Convert the input DEM to PCRaster if it isn’t already (i.e., a .map file).
if input_dem.lower().endswith('.map'):
feedback.pushInfo("Input DEM is already in PCRaster format.")
pcraster_dem = input_dem
else:
feedback.pushInfo("Converting input DEM to PCRaster format.")
pcraster_dem = os.path.join(temp_folder, "temp_dem.map")
self.ConvertToPCRaster(input_dem, pcraster_dem, gdalconst.GDT_Float32, "VS_SCALAR")
feedback.pushInfo("Reading DEM into memory.")
setclone(pcraster_dem)
dem_pcraster = readmap(pcraster_dem)
feedback.pushInfo("Generating water mask.")
watermask = pcrnot(defined(dem_pcraster))
water_mask_map = os.path.join(temp_folder, "temp_water_mask.map")
report(watermask, water_mask_map)
if sieve_threshold > 0:
feedback.pushInfo("Applying sieve filter.")
water_mask_sieved_tif = os.path.join(temp_folder, "temp_water_mask_sieved.tif")
self.sieve(water_mask_map, water_mask_sieved_tif, sieve_threshold)
water_mask_sieved_map = os.path.join(temp_folder, "temp_water_mask_sieved.map")
self.ConvertToPCRaster(water_mask_sieved_tif, water_mask_sieved_map, gdalconst.GDT_Byte, "VS_BOOLEAN")
feedback.pushInfo("Buffering water mask.")
watermask_sieved = readmap(water_mask_sieved_map)
setglobaloption("unitcell")
mask_buffer = spreadmaxzone(watermask_sieved, 0, 1, 2)
feedback.pushInfo("Segmenting water bodies.")
water_clumps = ifthenelse(mask_buffer == 1, clump(mask_buffer), nominal(mask_buffer))
feedback.pushInfo("Processing waterbody filling.")
cover_dsm = cover(dem_pcraster, 10000)
water_fill = ifthenelse(mask_buffer == 1, areaminimum(cover_dsm, water_clumps), dem_pcraster)
feedback.pushInfo("Saving output.")
final_output = output_filled
if os.path.exists(final_output):
try:
os.remove(final_output)
except Exception as e:
feedback.pushInfo("Warning: Could not remove existing file: " + str(e))
report(water_fill, final_output)
# Automatically load the final output into the QGIS map canvas.
from qgis.core import QgsProcessingContext
layer_details = QgsProcessingContext.LayerDetails(os.path.basename(final_output),
context.project(),
self.OUTPUT_FILLED)
context.addLayerToLoadOnCompletion(final_output, layer_details)
return {self.OUTPUT_FILLED: final_output}
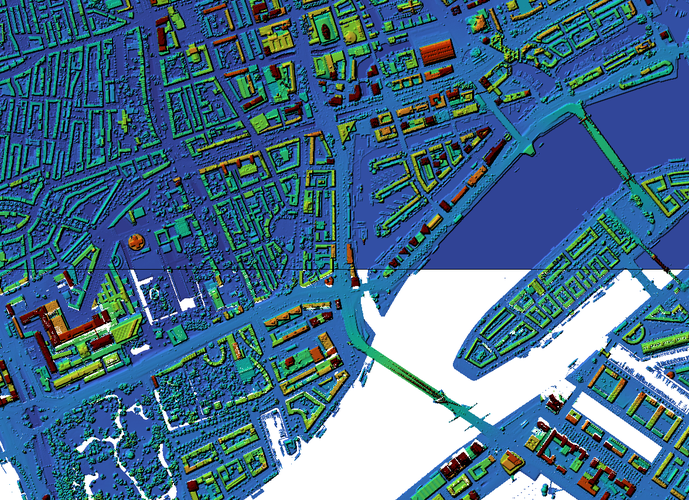
DEMs often have voids (nodata cells) for water bodies. This script is designed to address that issue by filling each water body with a uniform elevation value. The script first identifies nodata cells in the DEM. It then applies a 1-pixel buffer around these nodata cells. This buffer helps to capture the adjacent valid elevation data which represent the edges of the water bodies. For each water body, the script calculates the minimum elevation within the buffered area. This minimum value is assumed to be the most appropriate fill elevation for the given water body. Optionally, the result can be improved by applying a sieve filter.
Great, thank you very much!
Reviewed by gabrieldeluca 8 months, 1 week ago

This Processing Script is made available under the CC-0 license.